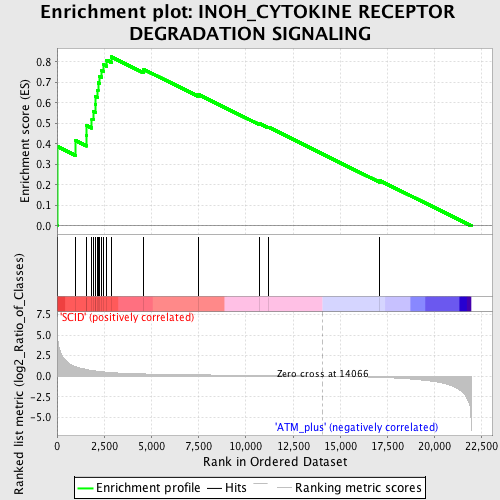
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | INOH_CYTOKINE RECEPTOR DEGRADATION SIGNALING |
| Enrichment Score (ES) | 0.82395643 |
| Normalized Enrichment Score (NES) | 1.8734571 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.005916808 |
| FWER p-Value | 0.058 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 8 | 6.245 | 0.3876 | Yes | ||
| 2 | PSMB9 | 1450696_at | 978 | 1.164 | 0.4157 | Yes | ||
| 3 | PSMB3 | 1417052_at | 1556 | 0.815 | 0.4399 | Yes | ||
| 4 | PSMA5 | 1424681_a_at 1434356_a_at | 1570 | 0.805 | 0.4894 | Yes | ||
| 5 | PSMC1 | 1416005_at | 1843 | 0.691 | 0.5199 | Yes | ||
| 6 | PSMA7 | 1423567_a_at 1423568_at | 1912 | 0.664 | 0.5580 | Yes | ||
| 7 | PSMB10 | 1448632_at | 2021 | 0.629 | 0.5921 | Yes | ||
| 8 | PSMB6 | 1448822_at | 2044 | 0.622 | 0.6298 | Yes | ||
| 9 | SOCS3 | 1416576_at 1455899_x_at 1456212_x_at | 2147 | 0.593 | 0.6619 | Yes | ||
| 10 | PSMA4 | 1460339_at | 2170 | 0.587 | 0.6974 | Yes | ||
| 11 | PSMB7 | 1416240_at | 2230 | 0.572 | 0.7302 | Yes | ||
| 12 | PSMB5 | 1415676_a_at | 2348 | 0.542 | 0.7586 | Yes | ||
| 13 | PSMB1 | 1420052_x_at 1420053_at 1448166_a_at | 2470 | 0.514 | 0.7850 | Yes | ||
| 14 | PSMA2 | 1448206_at | 2629 | 0.483 | 0.8078 | Yes | ||
| 15 | PSMB8 | 1422962_a_at 1444619_x_at | 2873 | 0.439 | 0.8240 | Yes | ||
| 16 | PSMD2 | 1415831_at | 4564 | 0.273 | 0.7638 | No | ||
| 17 | PSMA3 | 1448442_a_at | 7498 | 0.164 | 0.6401 | No | ||
| 18 | PSMB4 | 1438984_x_at 1451205_at | 10735 | 0.083 | 0.4977 | No | ||
| 19 | PSMB2 | 1444377_at 1448262_at 1459641_at | 11183 | 0.073 | 0.4818 | No | ||
| 20 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 17094 | -0.142 | 0.2209 | No |

